This repository is used to build the docker container used for CellOrganizer.
Installing Docker is beyond the scope of this document. To learn about Docker Community Edition (CE), click here.
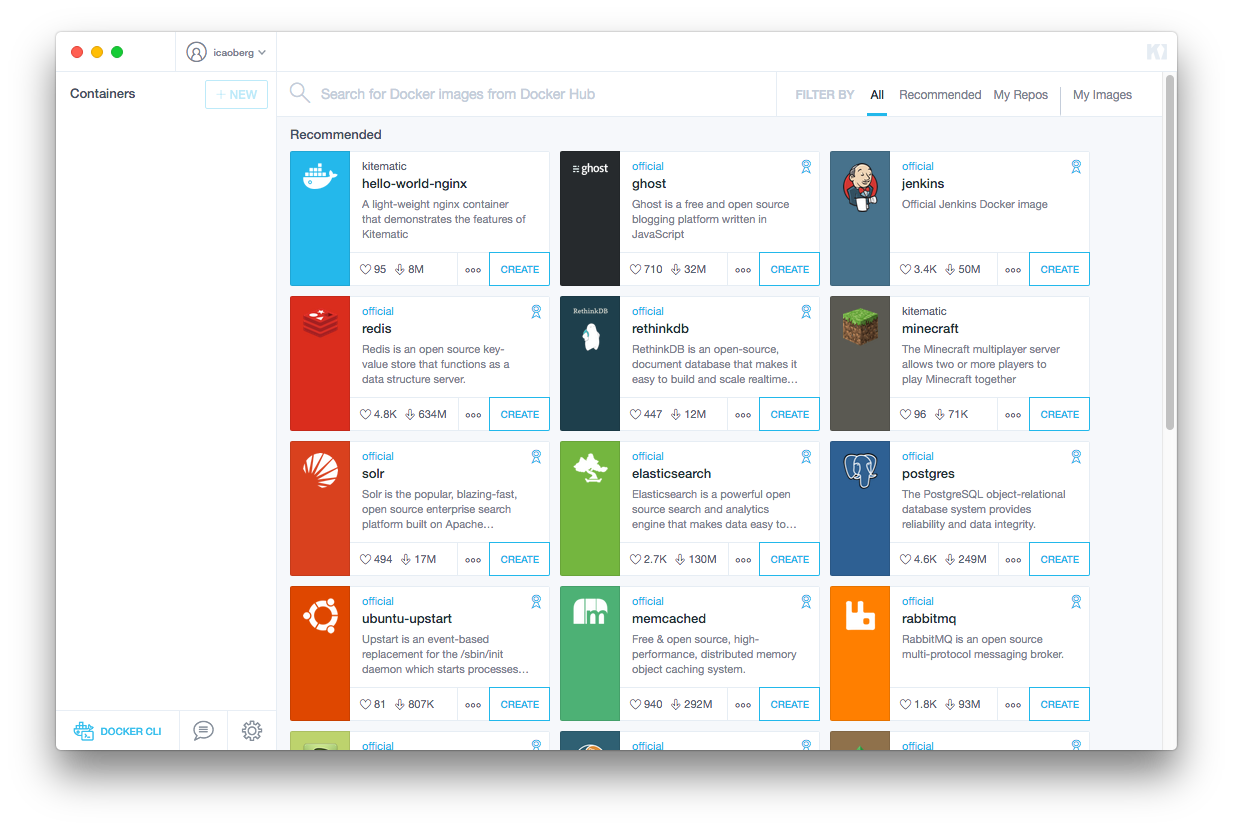
The easiest way to download an image and run a container is to use Kitematic.
- To install Kitematic, click here.
Running Kitematic will open a window that looks like this
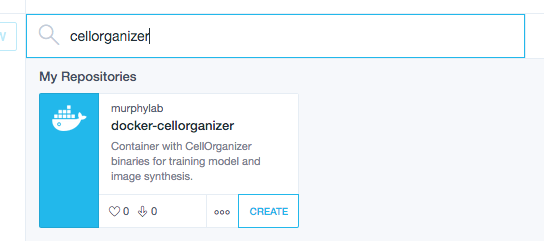
Use the searchbar to search for cellorganizer
and click Create.
To build an image using the Dockerfile in this repository, run the command
➜ docker build -t "murphylab/cellorganizer" .
The previous step should build an image
➜ docker container ls -a
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
48dde52f2bc8 murphylab/cellorganizer "/bin/bash -c 'pyt..." 45 seconds ago Exited (0) 39 seconds ago frosty_wescoff
To run a container using the image above
➜ docker run -i -t murphylab/cellorganizer
The container comes with
- CellOrganizer binaries
- Generative models
- Murphy Lab 2D/3D HeLa datasets
- BioFormats tools
- vim-as-an-IDE
When contributing to this repository, please first discuss the change you wish to make via issue, email, or any other method with the owners of this repository before making a change.
Support for CellOrganizer has been provided by grants GM075205, GM090033 and GM103712 from the National Institute of General Medical Sciences, grants MCB1121919 and MCB1121793 from the U.S. National Science Foundation, by a Forschungspreis from the Alexander von Humboldt Foundation, and by the Freiburg Institute for Advanced Studies.
Copyright (c) 2007-2019 by the Murphy Lab at the Computational Biology Department in Carnegie Mellon University