Neuron spike train and local field potential data decoding during experiment: http://www.jneurosci.org/content/36/5/1547
Previous Bayesian statistical model: https://www.ncbi.nlm.nih.gov/pubmed/28529731
Spike train data analysis: https://www.sciencedirect.com/science/article/pii/S0896627316302501
Convolutional neural networks: https://keras.io/getting-started/sequential-model-guide/#sequence-classification-with-1d-convolutions
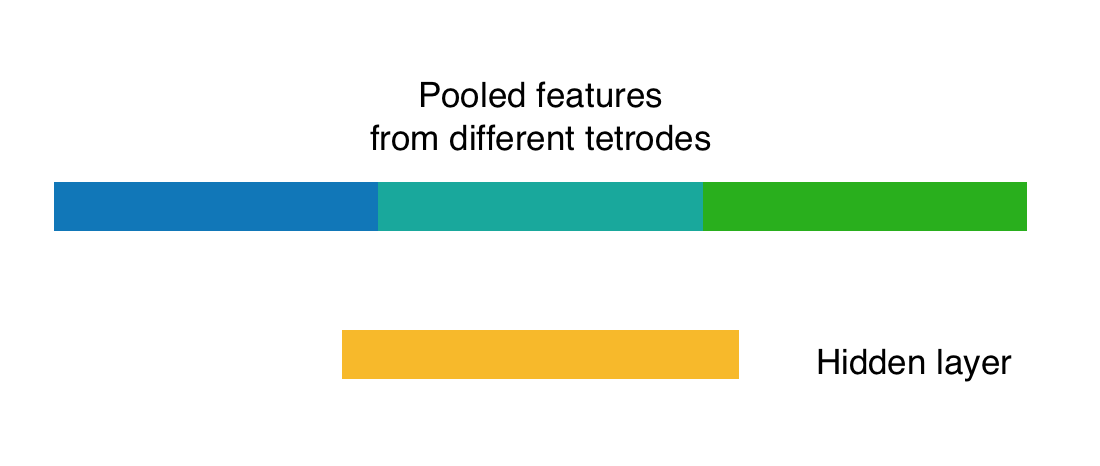
Tetrode-wise convolution: for each tetrode, convolution filters are applied on the multivariate time series of spike train and LFP data; pooled features from all the tetrodes are combined before a hidden layer and output layer.
data_utils.py process data arrays (trial_info, spike_data_binned, lfp_data_sampled) and organize data by tetrode
tetrode.py build neural network model in Keras and perform cross-validation during training
helper.py extract model hidden layer latent representation within different time windows
# process data
tetrode_data, decoding_target, tetrode_ids, tetrode_units, spike_data_binned, lfp_data_sampled = prepare_data(rat_name)
# build model
model = build_tetrode_model(tetrode_ids, tetrode_units)
# train model
hist = model.fit(tetrode_data, decoding_target, nb_epoch=100, batch_size=20, verbose=0, validation_split=0.1, shuffle=True)
# feature extraction
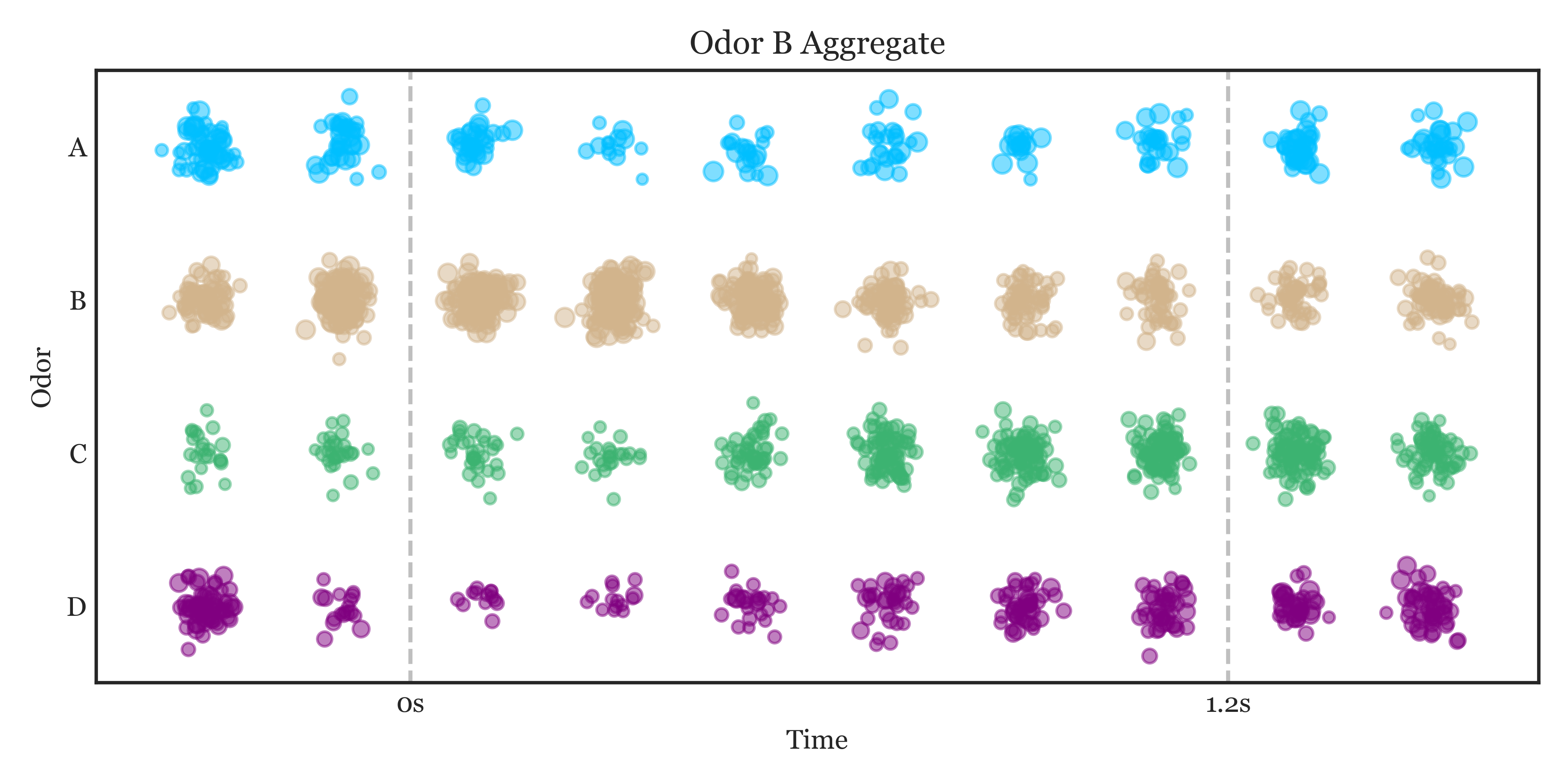
all_latent = extract_latent(intermediate_layer_model, spike_data_binned, lfp_data_sampled, tetrode_ids, tetrode_units, 25, 20)Result visualization: see notebooks tetrode_model.ipynb and aggregate_decoding.ipynb
Theta cycle replay analysis: theta cycles are determined with Hilbert transformation on filtered LFP data; spike train data are then aligned with theta phase rather than time.
data.py process data arrays (trial_info, spike_data_binned, lfp_data_sampled) and find reference LFP tetrode
theta.py calculate theta phase from LFP data and align spike train data
decoding.py train LASSO decoding model with cross-validation and get model prediciton
# perform theta analysis
rolling_preds_superchris, phase_preds_superchris, target_superchris = analysis_flow('superchris', theta_thres, corr_thres)
# theta phase prediction
decoding_preds1_superchris, decoding_preds2_superchris, decoding_preds3_superchris = phase_preds_superchris
# hypothesis test
p1 = paired_t_test(np.concatenate([decoding_preds1_B[:, 0], decoding_preds1_C[:, 1]]),
np.concatenate([decoding_preds1_B[:, 2], decoding_preds1_C[:, 3]]))Final results: see notebook final_theta_analysis.ipynb