-
Notifications
You must be signed in to change notification settings - Fork 1
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Units for trait evaluation observation values #13
Comments
|
thanks @jd-campbell. This may be something that we can fix when we can work from access to the underlying database, rather than the bulk exports that we have used in the past as our only mechanism. @sdash-github can probably confirm but I don't think that the bulk export files have the units. @ekcannon, if you have the db access now perhaps we can get your help on this someday. |
|
Right. the data didn't come with units when downloaded.
…On 2017/8/18 4:41 PM, adf-ncgr wrote:
thanks @jd-campbell <https://github.com/jd-campbell>. This may be
something that we can fix when we can work from access to the
underlying database, rather than the bulk exports that we have used in
the past as our only mechanism. @sdash-github
<https://github.com/sdash-github> can probably confirm but I don't
think that the bulk export files have the units. @ekcannon
<https://github.com/ekcannon>, if you have the db access now perhaps
we can get your help on this someday.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#13 (comment)>,
or mute the thread
<https://github.com/notifications/unsubscribe-auth/ADgOe3-0ZHG77KLjIT-oSGOBPPiOBgf8ks5sZhMKgaJpZM4O8GCs>.
|
|
If someone (@sdash-github?) gives me a list of desired information, I can attempt a query to create updated data (from December, 2016). I'll also request a new data dump from Pete Cyr. |
|
@ekcannon - to start with, we can share the files we have used for data import in the past. But I think that we'll need to spend some time looking over what all is available from the database before we can answer the question of how we'd like to augment the info set with units and whatever else might be in there. My recollection is that for most of the genera we had to make do with what we got directly downloaded from their website, but for Glycine we got files via David Grant that were a little differently structured and maybe had more "user friendly" descriptors associated with things- @sdash-github may recall better. Anyway, we'll get the files uploaded from our NCGR filesystem to where you can look at them... |
|
tarball now available on lis-* servers: ~adf/LIS_GERMPLASM.tgz can be rehomed to more appropriate place if there's one that makes sense. |
|
Could add a units column or combine with 'observation_value'. Also, the code values need to be displayed, e.g. SEEDPROD=4 is meaningless unless one knows that the value is on a 1-9 scale (though this example is too subjective to be of much use). Or that LYGUSBUG=3 means LYGUSBUG=3 leaves wilting. |
|
I think our current schema for the lis_germplasm.* tables is more or less just reflecting what is in those export files; my guess is that units and code values are probably described somewhere in the GRIN db in whatever is being referenced by the "method_name" column in the e.g. select * from lis_germplasm.legumes_grin_evaluation_data where method_name='S9.COWPEA.GH.1999'; so if we can get the GRIN info alluded to by the "method_name" we might just be able to add an extra table, and alter the display code to get the relevant metadata from there? hard to say exactly without more transparency into the GRIN db- will you be able to generate a schema diagram for it or is there one already extant somewhere? |
|
I have the schema in that I have access to the database, but haven't tried diagramming it (yet). Be warned that any given method_name may be linked to accessions for multiple 'crops'. For example, there is a barley method that is associated with a few maize observations. It looks like it may not be possible to extract specific unit information - have a question in to Pete - but instead it appears that units are only embedded in the trait description, e.g., "... measured in grams per ...." The descriptor data is loaded into Chado separately from the observations. How is this handled in the GIS tool? |
|
If I understand the question, I think the answer is "similarly separate", ie. which I believe are more or less loaded directly from those exported files, although I think some |
This suggestion came from the Germplasm short course I gave at ISU with GRIN-Global curator in the audience.
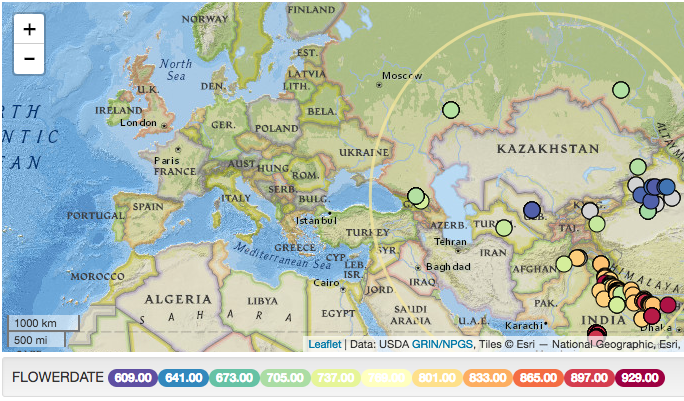
-- When looking at the trait data for a species there is no unit name, attached example FlowerDate in Glycine Max. Person in the class said trait data associated with GIS would be more helpful with units. The GRIN-Global curator said that the trait units is in the database.
The text was updated successfully, but these errors were encountered: