This module was designed to aid in collection and further analysis of raw phenotypic data. With this in mind it provides:
- An interface for defining projects including which traits are expected to be collected and the users collecting them;
- An interactive, researcher-friendly excel datasheet upload;
- Summary charts
- Data Download functionality with filter criteria and customizable R-friendly headers.
Note: there are some modifications needed to the spreadsheet reader to get it to handle dates the way we expect. Make sure to apply the spreadsheet-reader.patch included with this module to the spreadsheet-reader library.
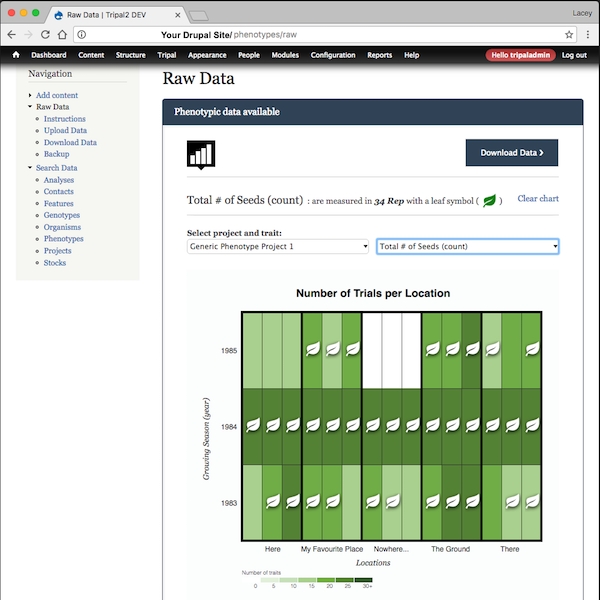
- A d3.js heatmap summarizes the raw data available by displaying the number of traits broken down by location (x-axis) and replicate (y-axis; grouped by year). This chart uses a materialized view for improved performance.
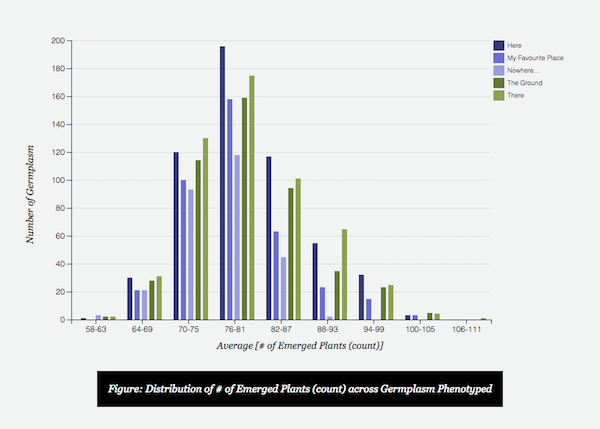
- If a trait is selected on the Heatmap summary, a barchart is dynamically generated beneath to show the range of values collected.
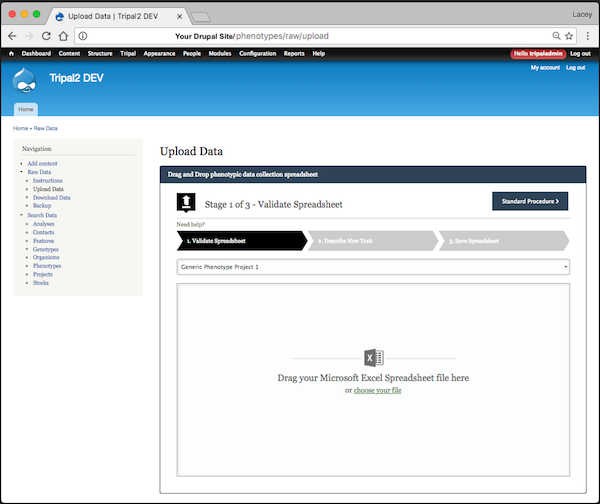
- Upload data functionality supporting excel spreadsheets (XLSX). The loader expects the traits specified for a given project but is also flexible enough to allow users to add additional traits (one per column) to the spreadsheet. If additional traits are present, the loader asks the user to describe the trait including the units and any scale used.
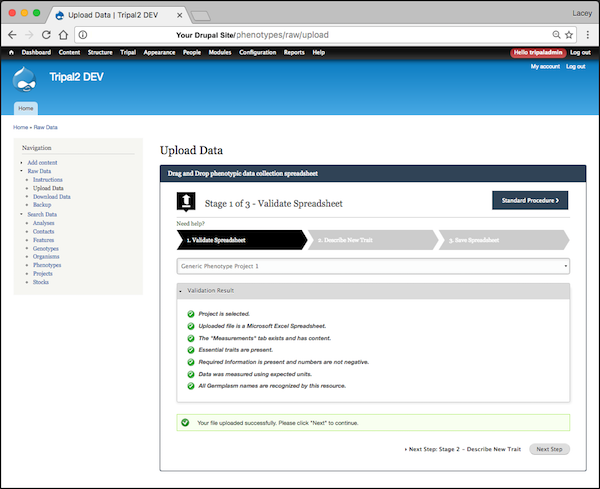
- During upload the file is validated using a number of tests including general "Is this an excel file?", as well as, "Do all the germplasm in the file already exist in the chado.stock table?". Validation tests are provided via a Drupal hook meaning you can add your own data-specific tests (see github wiki).
- A searchable trait collection instruction page defining a standard protocol for collection of the traits. This information is pulled from the trait controlled vocabulary and thus easily updatable.
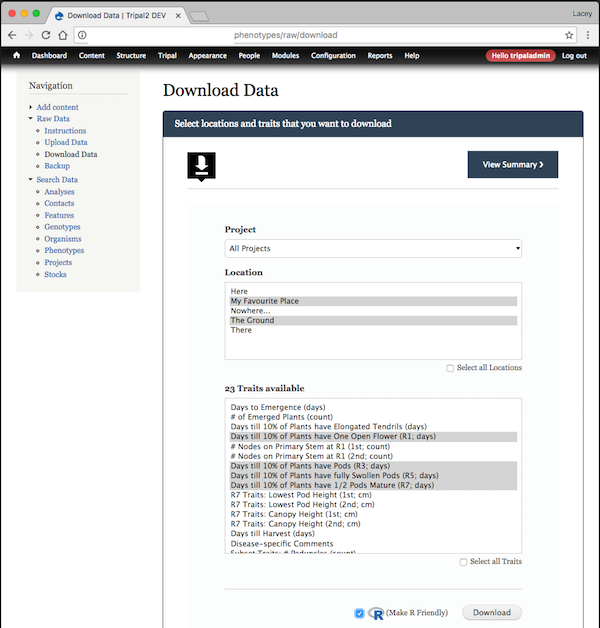
- Data Download functionality which allows users to select the locations and traits they are interested in. A comma-separated file is produced which can easily be opened in excel for viewing and is R-friendly for analysis.
Trait information is stored in a custom controlled vocabulary set-up by this module. All the remaining data is stored in a custom relational schema. This allows us to keep the raw data separate from chado and paves the way for an ND Phenotypes module which stored the analyzed/filtered phenotypic data in the chado schema and provides trait pages and summary information on germplasm and project pages.
In addition to this README there is a wiki available at https://github.com/UofS-Pulse-Binfo/rawphenotypes/wiki which includes an administration tutorial, as well as, other useful information.