class: middle, center, title-slide count: false
.huge.blue[Matthew Feickert]
.huge[(University of Wisconsin-Madison)]
[email protected]
September 18th, 2023
.kol-2-3[ .huge[
- Privileged opportunity to work among multiple scientific communities
- Care about .bold[reusable] open science to be able to push particle physics forward at the .bold[community scale]
- The challenges of the next decade provide wonderful research environments that will require interdisciplinary knowledge exchange to fully attack
- Today we'll share .bold[high level] views of deeply .bold[technical problems]
]
]
.kol-1-3[
.center.width-65[
 ]
]
.kol-1-2.center[
.caption[LHC] ] .kol-1-2.center[ .caption[ATLAS] ] .kol-1-1[ .kol-1-2.center[ ] .kol-1-2.center[ .kol-1-2.center[ ] .kol-1-2.center[ ] ] ].large[ * Increase in luminosity of roughly order of magnitude - $3$ - $4$ $\mathrm{ab}^{-1}$ (factor of 20-25 from Run-2 delivered) * Boon for measurements constrained by statistical uncertainties, searches for rare processes ]
.center.large[Challenge to be able to .bold[record, store, and analyze] the data]
.kol-1-2[
] .kol-1-2[]
.center.large[Projected .bold[required compute usage] for HL-LHC (want R&D below budget line)]
.center[ATLAS and CMS software and computing reviews]
.kol-1-2[
.huge[ * LHC experiments as stakeholders * LHC operations as partners ] ] .kol-1-2[ .caption[Institute for Research and Innovation in Software for High Energy Physics (IRIS-HEP)] ].kol-1-1[ .kol-1-2[ .huge[ Designed around focus areas ] .large[
- Intellectual Hub
- Analysis Systems
- Data Organization, Management, and Access (DOMA)
- Innovative Algorithms
- Translational Research for AI
- Scalable Systems Laboratory (SSL)
- OSG Services for LHC (OSG-LHC) ] ] .kol-1-2[
.caption[IRIS-HEP Institute Structure] ] ]
.large[ community engagement with .bold[training, education, and outreach] and .bold[institute grand challenges] ]
.huge[
- Deployable analysis pipelines that reduces physicist time-to-insight
- Tools integrate into the broader scientific Python computing ecosystem
- Analysis reuse as deployment feature ]
.huge[
- Integrating machine learning training and inference into analysis workflows
- c.f. Machine Learning for Columnar High Energy Physics Analysis, Elliott Kauffman, CHEP 2023 ]
.center.large[ In his PyCon 2017 keynote, Jake VanderPlas gave us the iconic "PyData ecosystem" image ]
.center.large[ In his 2022 PyHEP topical meeting update, Jim Pivarski gave us a view for the PyHEP ecosystem ]
.center.large["import XYZ" matches in GitHub repos for users who fork [CMSSW](https://github.com/cms-sw/cmssw) by file]
.footnote[Modern Python analysis ecosystem for High Energy Physics, Jim Pivarski, Matthew Feickert, Gordon Watts]
.center.large["import XYZ" matches in GitHub repos for users who fork [CMSSW](https://github.com/cms-sw/cmssw) by library/tool]
.footnote[Modern Python analysis ecosystem for High Energy Physics, Jim Pivarski, Matthew Feickert, Gordon Watts]
.center.large["pip install XYZ" download rate for MacOS/Windows (no batch jobs) in aggregate]
.footnote[Modern Python analysis ecosystem for High Energy Physics, Jim Pivarski, Matthew Feickert, Gordon Watts]
.center.large["pip install XYZ" download rate for MacOS/Windows (no batch jobs) by package] .caption[Aided by interoperable design]
.footnote[Modern Python analysis ecosystem for High Energy Physics, Jim Pivarski, Matthew Feickert, Gordon Watts]
.kol-1-1[ .kol-1-3[
] .kol-1-3[ ] .kol-1-3[ ] ] .kol-1-3[ .center.huge[[dask-awkward](https://github.com/dask-contrib/dask-awkward)].center[Native Dask collection for partioned Awkward arrays for analysis at scale] ] .kol-1-3[ .center.huge[scikit-build-core]
.center[Next generation of build tools for scientific packaging] ] .kol-1-3[ .center.huge[NumFOCUS]
.center[Organizing and supporting scientific open source] ]
.footnote[Taking a slide from Lukas Heinrich]
.kol-1-2[
] .kol-1-2.huge[.bold[New directions in science are launched by new tools much more often than by new concepts.]
— Freeman Dyson ]
- As we'll see later, having access to the gradient while performing minimization is highly beneficial!
- Can imagine multiple ways of arriving at gradients for computational functions
- But want them to be both .bold[exact] and .bold[flexible]
.center.width-25[ ]
.kol-6-8[
.bold.center[Symbolic]
.center.width-100[
]
.kol-6-8[
.bold.center[Symbolic]
.center.width-100[ ]
]
.kol-2-8.huge[
]
]
.kol-2-8.huge[
- Exact: .blue[Yes]
- Flexible: .red[No] ]
- As we'll see later, having access to the gradient while performing minimization is highly beneficial!
- Can imagine multiple ways of arriving at gradients for computational functions
- But want them to be both .bold[exact] and .bold[flexible]
.center.width-25[ ]
.kol-6-8[
.bold.center[Numeric]
.center.width-70[
]
.kol-6-8[
.bold.center[Numeric]
.center.width-70[ ]
]
.kol-2-8.huge[
]
]
.kol-2-8.huge[
- Exact: .red[No]
- Flexible: .blue[Yes] ]
- As we'll see later, having access to the gradient while performing minimization is highly beneficial!
- Can imagine multiple ways of arriving at gradients for computational functions
- But want them to be both .bold[exact] and .bold[flexible]
.center.width-25[ ]
.kol-6-8[
.bold.center[Automatic]
.center.width-80[
]
.kol-6-8[
.bold.center[Automatic]
.center.width-80[ ]
]
.kol-2-8.huge[
]
]
.kol-2-8.huge[
- Exact: .blue[Yes]
- Flexible: .blue[Yes] ]
.kol-3-5[
- Automatic differentiation (autodiff) provides gradients of numerical functions to machine precision
- Build computational graph of the calculation
- Nodes represent operations, edges represent flow of gradients
- Apply the chain rule to operations
- Can traverse the graph in forward or reverse modes depending on the relative dimensions of input and output for efficient computation
]
.kol-2-5.center[
.width-100[ ]
]
]
]
.grid[ .kol-1-2.large[
- Allows writing fully differentiable programs that are efficient and accurate
- Resulting system can be optimized end-to-end using efficient gradient-based optimization algorithms
- Exploit advances in deep learning
- Enables .italic[efficient] computation of gradients and Jacobians
- Large benefit to statistical inference
- Replace non-differentiable operations with differentiable analogues
- Binning, sorting, cuts
]
.kol-1-2[
.center.width-100[ ]
.center[Snowmass 2021 LOI]
]
]
]
.center[Snowmass 2021 LOI]
]
]
- Binning, sorting, cuts
]
.kol-1-2[
class: focus-slide, center
.huge.bold.center[Application of automatic differentiation in pyhf]
.kol-1-1[
.kol-1-3.center[
.width-100[ ]
Search for new physics
]
.kol-1-3.center[
]
Search for new physics
]
.kol-1-3.center[
.width-100[ ]
]
Make precision measurements ] .kol-1-3.center[ .width-110[[](https://atlas.web.cern.ch/Atlas/GROUPS/PHYSICS/PAPERS/SUSY-2018-31/)]
Provide constraints on models through setting best limits ] ]
- All require .bold[building statistical models] and .bold[fitting models] to data to perform statistical inference
- Model complexity can be huge for complicated searches
- Problem: Time to fit can be .bold[many hours]
- .blue[Goal:] Empower analysts with fast fits and expressive models
- A flexible probability density function (p.d.f.) template to build statistical models in high energy physics
- Developed in 2011 during work that lead to the Higgs discovery [CERN-OPEN-2012-016]
- Widely used by ATLAS for .bold[measurements of known physics] (Standard Model) and .bold[searches for new physics] (beyond the Standard Model)
.kol-2-5.center[
.width-90[ ]
.bold[Standard Model]
]
.kol-3-5.center[
.width-100[
]
.bold[Standard Model]
]
.kol-3-5.center[
.width-100[ ]
.bold[Beyond the Standard Model]
]
]
.bold[Beyond the Standard Model]
]
.center[$\textcolor{#00a620}{\vec{n}}$: .obsdata[events],
.bold[Use:] Multiple disjoint channels (or regions) of binned distributions with multiple samples contributing to each with additional (possibly shared) systematics between sample estimates
.bold[Main pieces:]
- .blue[Main Poisson p.d.f. for simultaneous measurement of multiple channels]
- .katex[Event rates]
$\nu_{cb}(\textcolor{#0495fc}{\vec{\eta}}, \textcolor{#9c2cfc}{\vec{\chi}})$ (nominal rate$\nu_{scb}^{0}$ with rate modifiers)- encode systematic uncertainties (e.g. normalization, shape)
- .red[Constraint p.d.f. (+ data) for "auxiliary measurements"]
.center[$\textcolor{#00a620}{\vec{n}}$: .obsdata[events],
.bold[Use:] Multiple disjoint channels (or regions) of binned distributions with multiple samples contributing to each with additional (possibly shared) systematics between sample estimates
.bold[Main pieces:]
- .blue[Main Poisson p.d.f. for simultaneous measurement of multiple channels]
- .katex[Event rates]
$\nu_{cb}(\textcolor{#0495fc}{\vec{\eta}}, \textcolor{#9c2cfc}{\vec{\chi}})$ (nominal rate$\nu_{scb}^{0}$ with rate modifiers)- encode systematic uncertainties (e.g. normalization, shape)
- .red[Constraint p.d.f. (+ data) for "auxiliary measurements"]
Mathematical grammar for a simultaneous fit with:
- .blue[multiple "channels"] (analysis regions, (stacks of) histograms) that can have multiple bins
- with systematic uncertainties that modify the event rate
$\nu_{cb}(\textcolor{#0495fc}{\vec{\eta}}, \textcolor{#9c2cfc}{\vec{\chi}})$ - coupled to a set of .red[constraint terms]
.center.width-40[ ]
.center[Example: .bold[Each bin] is separate (1-bin) channel, each .bold[histogram] (color)
]
.center[Example: .bold[Each bin] is separate (1-bin) channel, each .bold[histogram] (color)
is a sample and share a .bold[normalization systematic] uncertainty]
.center[$\textcolor{#00a620}{\vec{n}}$: .obsdata[events],
.center[.bold[This is a mathematical representation!] Nowhere is any software spec defined]
.center[.bold[Until 2018] the only implementation of HistFactory has been in ROOT]
.kol-1-2.large[
- First non-ROOT implementation of the HistFactory p.d.f. template
- pure-Python library as second implementation of HistFactory
$ python -m pip install pyhf- No dependence on ROOT!
.center.width-100[ ]
]
.kol-1-2.large[
]
]
.kol-1-2.large[
- Open source tool for all of HEP
- IRIS-HEP supported Scikit-HEP project
- Used in ATLAS SUSY, Exotics, and Top groups in 25 published analyses (inference and published models)
- Used by Belle II
(DOI: 10.1103/PhysRevLett.127.181802) and MicroBooNE (upcoming results) - Used in analyses and for reinterpretation by phenomenology community,
SModelS
(DOI: 10.1016/j.cpc.2021.107909), andMadAnalysis 5(arXiv:2206.14870) - Maybe your experiment too! ]
.grid[ .kol-2-3[
- All numerical operations implemented in .bold[tensor backends] through an API of
$n$ -dimensional array operations - Using deep learning frameworks as computational backends allows for .bold[exploitation of auto differentiation (autodiff) and GPU acceleration]
- As huge buy in from industry we benefit for free as these frameworks are .bold[continually improved] by professional software engineers (physicists are not)
.kol-1-2.center[
.width-80[ ]
]
.kol-1-2[
]
]
.kol-1-2[
- Hardware acceleration giving .bold[order of magnitude speedup] in interpolation for systematics!
- does suffer some overhead
- Noticeable impact for large and complex models
.width-50[] ] ]
With tensor library backends gain access to exact (higher order) derivatives — accuracy is only limited by floating point precision
.grid[ .kol-1-2[ .large[Exploit .bold[full gradient of the likelihood] with .bold[modern optimizers] to help speedup fit!]
.large[Gain this through the frameworks creating computational directed acyclic graphs and then applying the chain rule (to the operations)]
]
.kol-1-2[
.center.width-80[ ]
]
]
]
]
]
.footnote[Example adapted from Lukas Heinrich's PyHEP 2020 tutorial]
.kol-1-2.center[
] .kol-1-2.center[ ].bold.center[Having access to the gradients can make the fit orders of magnitude faster than finite difference]
.footnote[Example adapted from Lukas Heinrich's PyHEP 2020 tutorial]
.kol-1-2.center[
] .kol-1-2.center[ ].bold.center[Having access to the gradients can make the fit orders of magnitude faster than finite difference]
class: focus-slide, center
.huge.bold.center[Familiar (toy) example: Optimizing selection "cut" for an analysis]
- Counting experiment for presence of signal process
- Place discriminate selection cut on observable
$x$ to maximize significance- Significance:
$\sqrt{2 (S+B) \log(1 + \frac{S}{B})-2S}$ (for small$S/B$ : significance$\to S/\sqrt{B}$ )
- Significance:
.footnote[Example inspired by Alexander Held's example of a differentiable analysis]
.kol-1-2.center[
] .kol-1-2.center[ ]- Set baseline cut at
$x=0$ (accept everything) - Step along cut values in
$x$ and calculate significance at each cut. Keep maximum.
.kol-1-2.center[
.width-100[ ]
]
.kol-1-2[
.width-100[
]
]
.kol-1-2[
.width-100[ ]
]
]
]
.center[Significance:
.kol-1-2.large[
- Need differentiable analogue to non-differentiable cut
- Weight events using activation function of sigmoid
.center[$w=\left(1 + e^{-\alpha(x-c)}\right)^{-1}$]
- Event far .italic[below] cut:
$w \to 0$ - Event far .italic[above] cut:
$w \to 1$ -
$\alpha$ tunable parameter for steepness- Larger
$\alpha$ more cut-like ] .kol-1-2[
- Larger
.width-100[] ]
.kol-1-2.large[
- For hard cuts the significance was calculated by applying the cut and than using the remaining
$S$ and$B$ events - But for the differentiable model there aren't cuts, so approximate cuts with the sigmoid approach and weights
- Comparing the two methods shows good agreement
- Can see that the approximation to the hard cuts improves with larger
$\alpha$ - But can become unstable, so tunable ] .kol-1-2.center[
.width-100[] ]
.kol-1-2.large[
- For hard cuts the significance was calculated by applying the cut and then using the remaining
$S$ and$B$ events - But for the differentiable model there aren't cuts, so approximate cuts with the sigmoid approach and weights
- Comparing the two methods shows good agreement
- Can see that the approximation to the hard cuts improves with larger
$\alpha$ - But can become unstable, so tunable ] .kol-1-2.center[
.width-100[] ]
.kol-2-5.large[
- Most importantly though, with the differentiable model we have access to the gradient
$\partial_{x} f(x)$
- So can find the maximum significance at the point where the gradient of the significance is zero
$\partial_{x} f(x) = 0$
- With the gradient in hand this cries out for automated optimization! ] .kol-3-5.center[
.kol-2-5.large[
- With a simple gradient descent algorithm can easily automate the significance optimization
- For this toy example, obviously less efficient then cut and count scan
- Gradient methods apply well in higher dimensional problems
- Allows for the "cut" to become a parameter that can be differentiated through for the larger analysis
]
.kol-3-5.center[
.width-100[
 ]
]
]
.kol-1-2[
- Provide differentiable analogue to histograms with kernel density estimation (KDE) or softmax
- Need smooth change compared to abrupt changes in binned yields
- Samples fed into NN that produces observable (NN output) KDE transformed and histogrammed.
- Construct
pyhfmodel with observable and perform inference to get$\mathrm{CL}_{s}$ for POI. - Backpropagate the
$\mathrm{CL}_{s}$ to update weights for NN.
.center.width-40[[](https://github.com/gradhep/neos)] .footnote[Graphics from [Nathan Simpson's PyHEP 2020 talk](https://indico.cern.ch/event/882824/timetable/#46-neos-physics-analysis-as-a)] ] .kol-1-2.center[ .width-40[[](https://indico.cern.ch/event/882824/timetable/#46-neos-physics-analysis-as-a)] .width-100[[](https://indico.cern.ch/event/882824/timetable/#46-neos-physics-analysis-as-a)] ]
.center[neos 3 bin KDE transformed observable (NN output) optimized with systematics w.r.t. 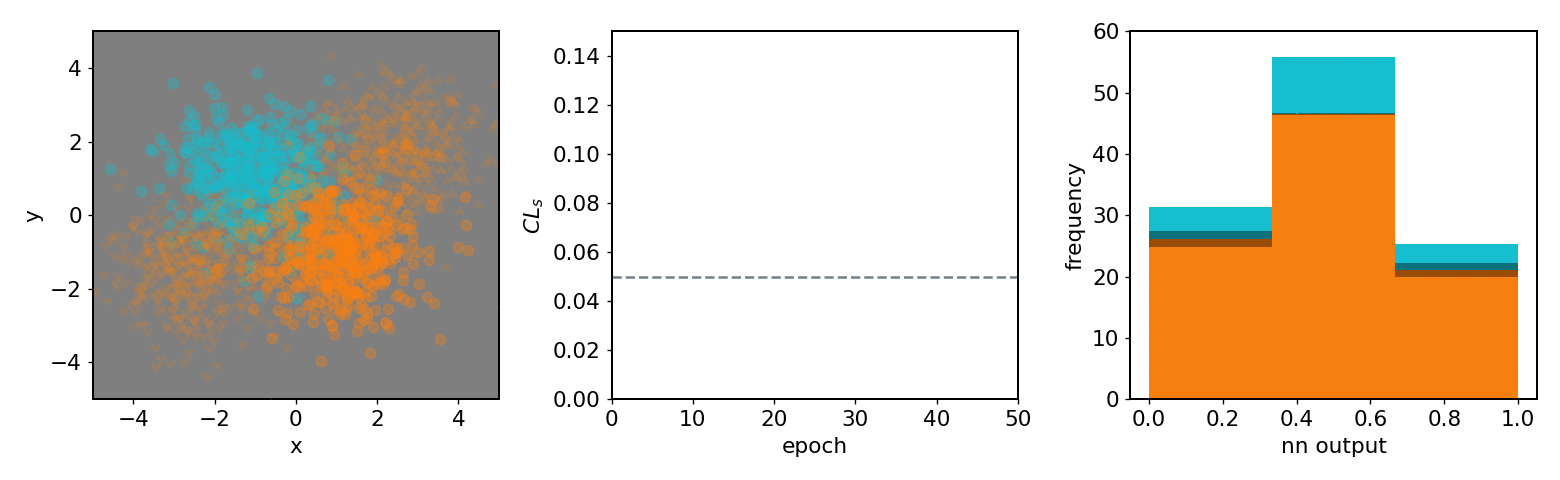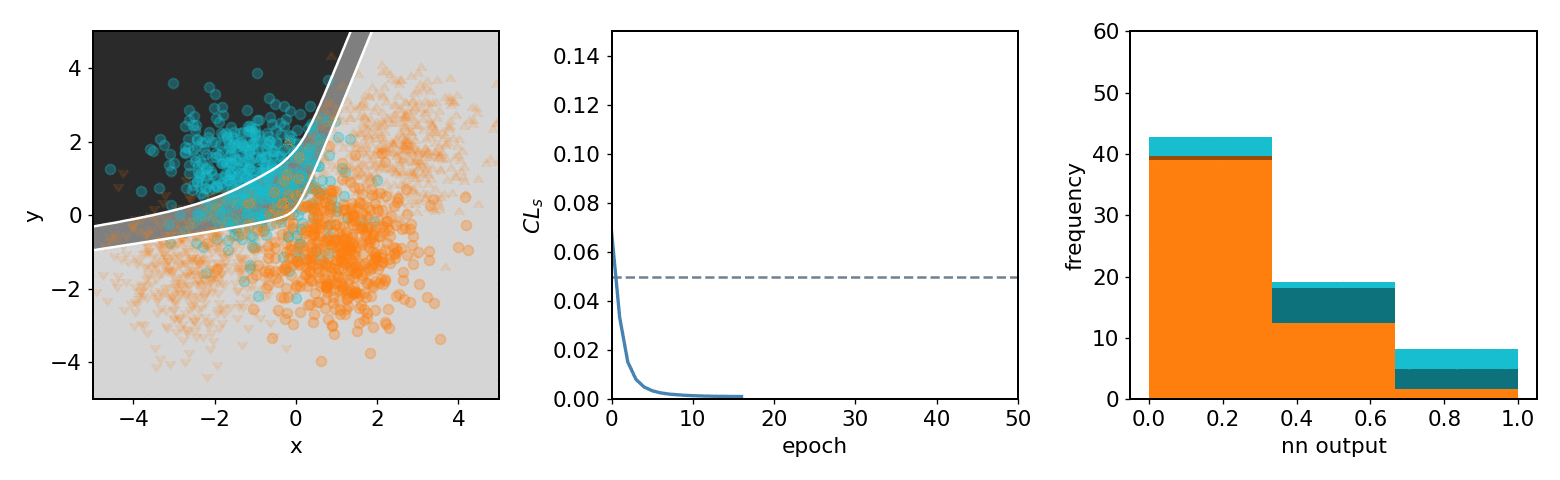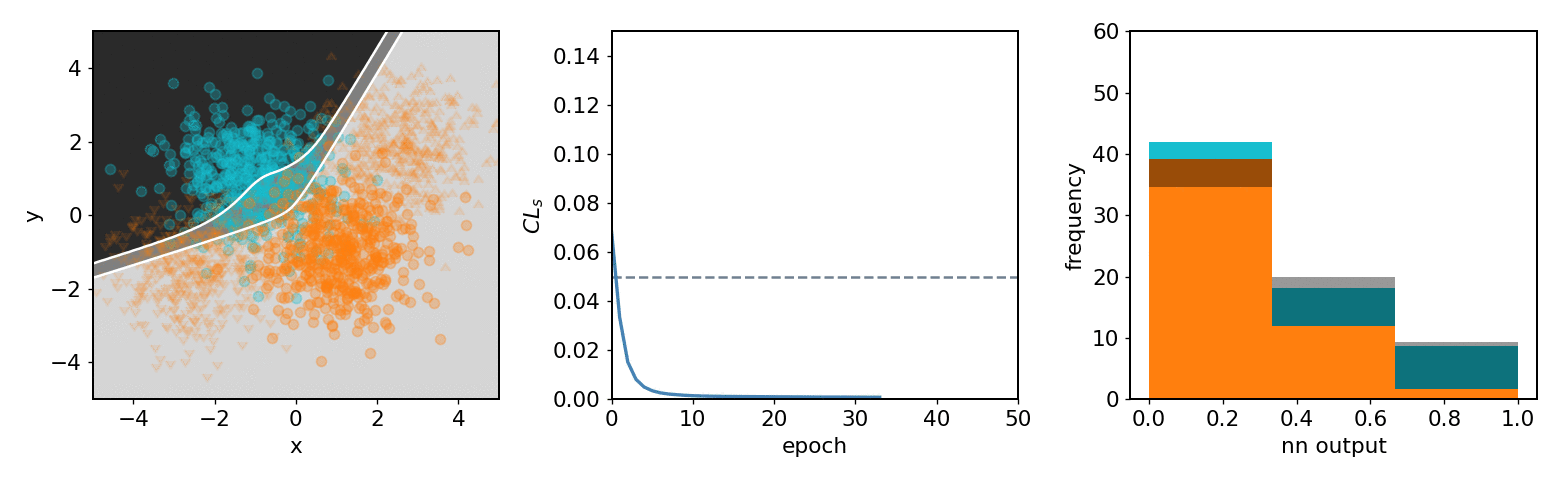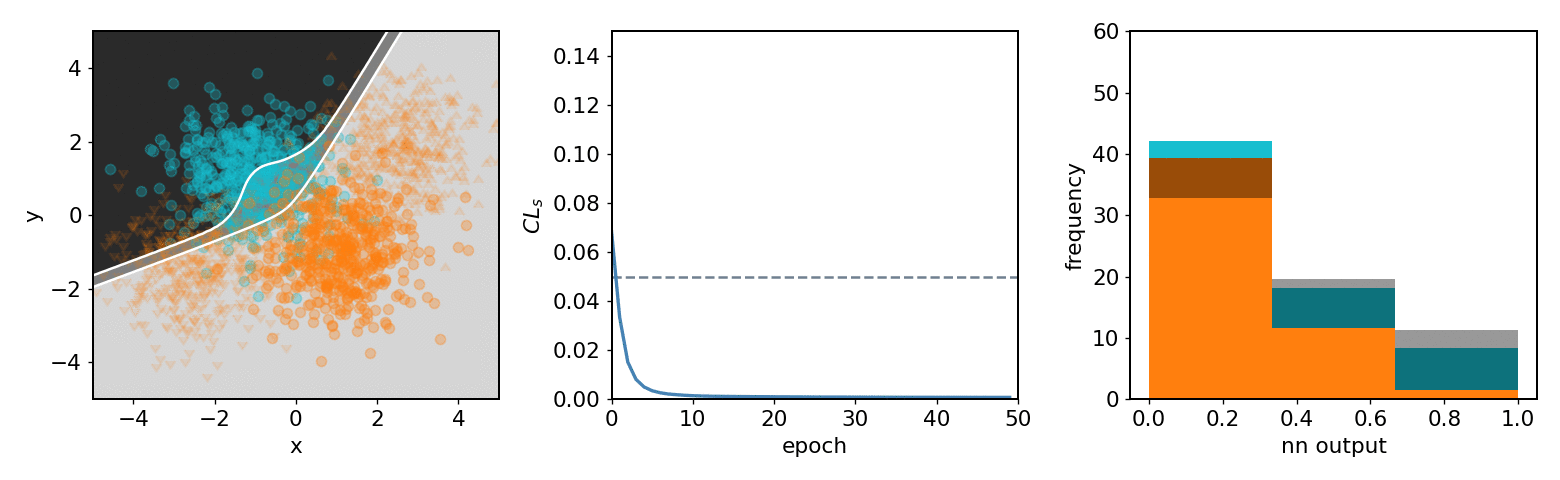 ]
]
.kol-1-3[
- .neos-orange[Background] and .neos-blue[signal] samples
- Same colors for dist. / hist.
- 3 decision regions are mappings of NN output
-
$[0.67, 1.0]$ bin$\to$ top left region ] .kol-1-3[
-
- From KDE of NN output form
pyhfmodel with 1 channel with 2 samples and 3 bins -
$\mathrm{CL}_{s}$ value minimized as goal of NN ] .kol-1-3[ - Observations in NN output
-
$0$ : Background-like -
$1$ : Signal-like
-
- Binned contents channel input for
pyhfmodel ]
class: focus-slide, center
.huge.bold.center[Differentiable analyses at LHC scale]
From the 2023 MIAPbP Workshop on on Differentiable and Probabilistic Programming for physics engagement with the broader community showed multiple large scale workflows
.center[.bold[If] things are differentiable, shouldn't be scared of .bold[large-scale codebases and applications]]
.kol-1-2[
] .kol-1-2[ .center[[Nicolas Gauger, MIAPbP Workshop 2023](https://indico.ph.tum.de/event/7314/contributions/7432/)] ].kol-2-5.code-large[
- Real world high energy physics analyses have various challenges:
- Computations highly complex chains
- Not implementable in a single framework
- Asynchronous multi-step procedures
- Strong need for distributed computing
- Passing of gradients .bold[between] different implementations and services
- Large scale machine learning in industry needs to do this to train models
- Possible solution to allow for distributed computations at scale exploiting gradients ] .kol-3-5.center[
.width-100[[](https://indico.cern.ch/event/960587/contributions/4070325/)] .caption[[Differentiating through PyTorch, JAX, and TensorFlow using FaaS](https://indico.cern.ch/event/960587/contributions/4070325/), Lukas Heinrich] ]
.center[Revisiting IRIS-HEP Analysis Systems in the context of distributed scaling and analysis reuse]
.large[
- Data and analyses done at the LHC are unique physics opportunities
- RECAST has been implemented in ATLAS as an enabling technology
- Resulting in ATLAS PUB notes extending the physics reach of original publications ]
.kol-1-3[
.caption[[ATL-PHYS-PUB-2019-032](https://inspirehep.net/literature/1795215)] ] .kol-1-3[ .caption[[ATL-PHYS-PUB-2020-007](https://inspirehep.net/literature/1795203)] ] .kol-1-3[ .caption[[ATL-PHYS-PUB-2021-020](https://inspirehep.net/literature/1870397)] ].kol-1-2[ .huge[ Leveraging REANA reproducible research data analysis platform possible to run distributed ML and analysis workflows at scale ]
.caption[[ Christian Weber, Reinterpretation Forum 2023](https://conference.ippp.dur.ac.uk/event/1178/contributions/6449/)] ] .kol-1-2[ .caption[[ATL-PHYS-PUB-2023-010](https://atlas.web.cern.ch/Atlas/GROUPS/PHYSICS/PUBNOTES/ATL-PHYS-PUB-2023-010/)] ].huge[
- General techniques and technologies applied to HEP problems, but not constrained to them
- Automatic differentiation is a rich field of research unto itself
- Engagement with the broader scientific open source community
- Planning for analysis reuse brings flexibility to leverage tooling ]
.huge[
- Many challenges and opportunities ahead at the HL-LHC
- Engaging the broader scientific open source community has been a boon for particle physics tooling
- Automatic differentiation gives a powerful tool in the form of differentiable programming
- Scalable and reusable analysis workflows allow leveraging our tools ]
class: end-slide, center
.large[Backup]
.center.large[Challenge to be able to .bold[record, store, and analyze] the data]
.kol-1-2[
] .kol-1-2[]
.center.large[Projected .bold[required disk usage] for HL-LHC (want R&D below budget line)]
.center[ATLAS and CMS software and computing reviews]
.center[Performing maps
.center[aka, "wide" vs. "tall" transformations]
.kol-1-2[
- .bold[Forward] mode
- Column wise evaluation of Jacobian
- Jacobian-vector products
- Execution time scales with input parameters
- Example: few variables into very high dimensional spaces
$\mathbb{R} \to \mathbb{R}^{100}$ ] .kol-1-2[
- .bold[Reverse] mode
- Row wise evaluation of Jacobian
- vector-Jacobian products
- Execution time scales with output parameters
- Example: scalar maps from very high-dimensional spaces
$\mathbb{R}^{100} \to \mathbb{R}$ ]
.center[Allows for efficient computation depending on dimensionality]
.bold[Use:] Multiple disjoint channels (or regions) of binned distributions with multiple samples contributing to each with additional (possibly shared) systematics between sample estimates
.kol-1-2[ .bold[Main pieces:]
- .blue[Main Poisson p.d.f. for simultaneous measurement of multiple channels]
- .katex[Event rates]
$\nu_{cb}$ (nominal rate$\nu_{scb}^{0}$ with rate modifiers) - .red[Constraint p.d.f. (+ data) for "auxiliary measurements"]
- encode systematic uncertainties (e.g. normalization, shape)
-
$\vec{n}$ : events,$\vec{a}$ : auxiliary data,$\vec{\eta}$ : unconstrained pars,$\vec{\chi}$ : constrained pars ] .kol-1-2[ .center.width-100[ ]
.center[Example: .bold[Each bin] is separate (1-bin) channel,
]
.center[Example: .bold[Each bin] is separate (1-bin) channel,
each .bold[histogram] (color) is a sample and share
a .bold[normalization systematic] uncertainty] ]
Mathematical grammar for a simultaneous fit with
- .blue[multiple "channels"] (analysis regions, (stacks of) histograms)
- each region can have .blue[multiple bins]
- coupled to a set of .red[constraint terms]
.center[.bold[This is a mathematical representation!] Nowhere is any software spec defined]
.center[.bold[Until recently] (2018), the only implementation of HistFactory has been in ROOT]
.bold[pyhf: HistFactory in pure Python]
.center.width-40[ ]
]
.kol-4-7[
- In HEP common for systematic uncertainties to be specified with two template histograms: "up" and "down" variation for parameter
$\theta \in \{\textcolor{#0495fc}{\vec{\eta}}, \textcolor{#9c2cfc}{\vec{\chi}} \}$ - "up" variation: model prediction for
$\theta = +1$ - "down" variation: model prediction for
$\theta = -1$ - Interpolation and extrapolation choices provide .bold[model predictions
$\nu(\vec{\theta},)$ for any$\vec{\theta}$ ]
- "up" variation: model prediction for
-
Constraint terms
$c_{j} \left(\textcolor{#a3130f}{a_{j}}\middle|\textcolor{#9c2cfc}{\theta_{j}}\right)$ used to model auxiliary measurements. Example for Normal (most common case):- Mean of nuisance parameter
$\textcolor{#9c2cfc}{\theta_{j}}$ with normalized width ($\sigma=1$ ) - Normal: auxiliary data
$\textcolor{#a3130f}{a_{j} = 0}$ (aux data function of modifier type) - Constraint term produces penalty in likelihood for pulling
$\textcolor{#9c2cfc}{\theta_{j}}$ away from auxiliary measurement value - As
$\nu(\vec{\theta},)$ constraint terms inform rate modifiers (.bold[systematic uncertainties]) during simultaneous fit - Example: Correlated shape
histosysmodifier could represent part of the uncertainty associated with a jet energy scale ] .kol-3-7[ .center.width-70[ ]
.center[Image credit: Alex Held]
]
]
.center[Image credit: Alex Held]
]
- Mean of nuisance parameter
Please checkout the many resources we have starting with the website and the SciPy 2020 talk!
.grid[ .kol-1-3.center[ .width-60[[](https://scikit-hep.org/)] ] .kol-1-3.center[.width-60[[](https://github.com/scikit-hep/pyhf)] ] .kol-1-3.center[
.width-70[[](https://iris-hep.org/)] ] ]
gradhep
]
.kol-1-3.center[
.width-100[ ]
]
neos, INFERNO
]
.kol-1-3.center[
.width-100[ ]
]
ACTS
]
.kol-1-1[ .bold.center[Groups, libraries, and applications growing rapidly] ]
- Lukas Heinrich, .italic[Distributed Gradients for Differentiable Analysis], Future Analysis Systems and Facilities Workshop, 2020.
- Jim Pivarski, .italic[History and Adoption of Programming Languages in NHEP], Software & Computing Round Table, 2022.
class: end-slide, center count: false
The end.

































