class: middle, center, title-slide count: false
KyungEon Choi, .blue[Matthew Feickert], Nikolai Hartmann, Lukas Heinrich, Alexander Held, Evangelos Kourlitis,
Nils Krumnack, Giordon Stark, Matthias Vigl, Gordon Watts on behalf of the .bold[ATLAS Computing Activity]
.large[(University of Wisconsin-Madison)]
[email protected]
International Conference on Computing in High Energy and Nuclear Physics (CHEP) 2024
October 21st, 2024
.middle-logo[]
.kol-1-2[
.caption[([ATLAS Software and Computing HL-LHC Roadmap](https://cds.cern.ch/record/2802918), 2022)] .large[ * Won't be able to store everything on disk * Move towards "trade disk for CPU" model ] ] .kol-1-2[ .caption[(Jana Schaarschmidt, [CHEP 2023](https://indico.jlab.org/event/459/contributions/11586/))]- Common file format for .bold[Run 4 Analysis Model]
- Monolithic: Intended to serve ~80% of physics analysis in Run 4
- Contains already-calibrated objects for fast analysis
- Will be able to use directly without need for ntuples ]
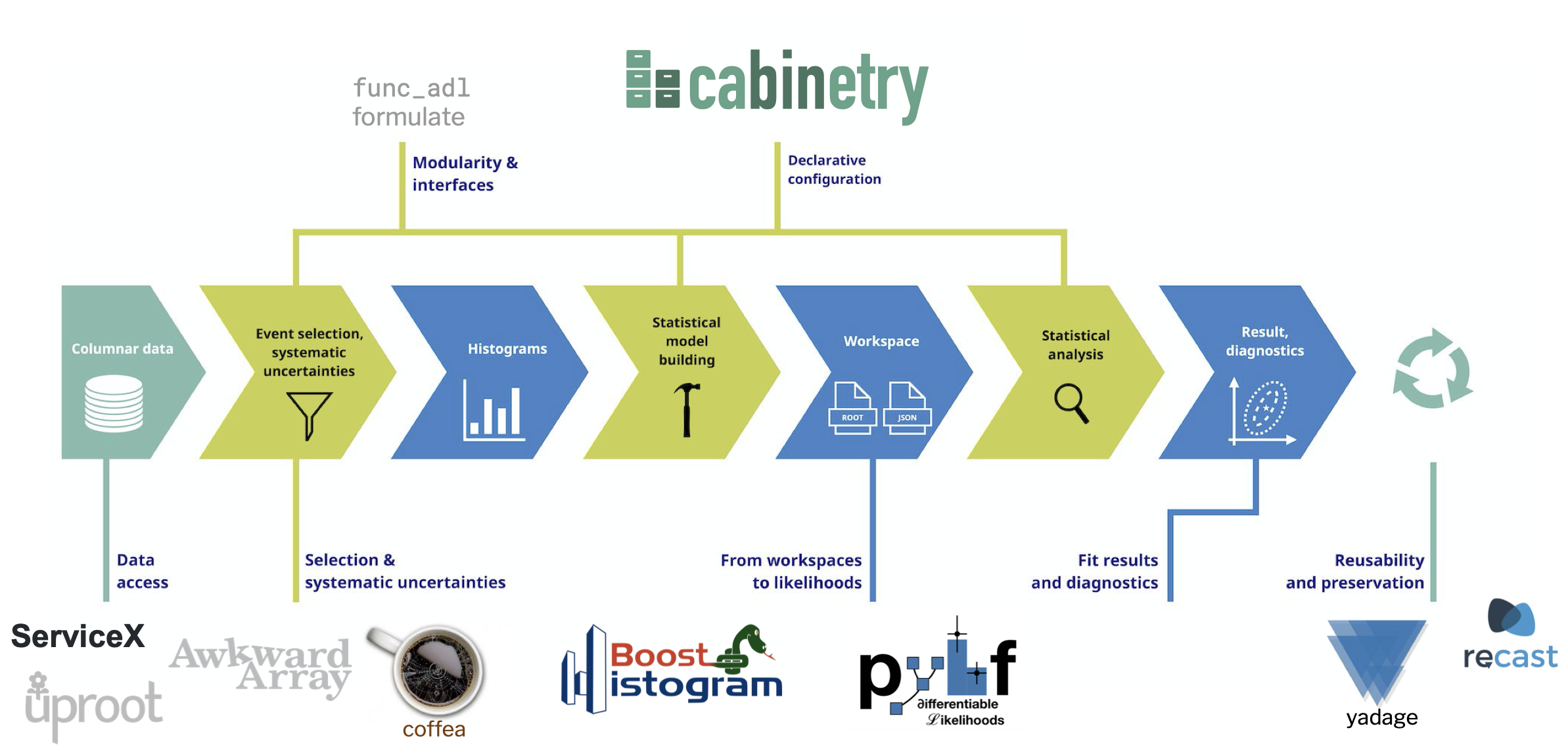
.kol-1-3[ .large[ Providing the elements of a .bold[columnar analysis pipeline]
- Data query and access
- Reading data files (ROOT and others) and columnar access
- Data transformation and histogramming
- Distributed analysis frameworks
- Statistical inference
- Analysis reinterpretation ] ] .kol-2-3[
]
.kol-1-5[
End user analysis ideally uses .bold[smaller and calibrated PHYSLITE]
.bold[Can still use PHYS] (same data format) through will need to perform .bold[additional steps] (calibration) with funcADL
]
.kol-4-5[
 .center.large[Components of an ATLAS [Analysis Grand Challenge (AGC)](https://agc.readthedocs.io/)
.center.large[Components of an ATLAS [Analysis Grand Challenge (AGC)](https://agc.readthedocs.io/)
demonstrator pipeline .smaller[(c.f. [The 200Gbps Challenge](https://indico.cern.ch/event/1338689/contributions/6009824/) (Alexander Held, Monday plenary))]]
.kol-1-2[ .large[
- Raw PHYSLITE is not easily loadable by columnar analysis tools outside of ROOT
- Challenges for correctly handling
ElementLinksand custom objects .smaller[(e.g. triggers)]
- Challenges for correctly handling
- Awkward Array supports
behaviors, which allow for efficiently reinterpreting data on the fly - ATLAS members have contributed to open ecosystem development to support PHYSLITE in both Uproot and Coffea
- Continuing to support fixes to both the PyHEP ecosystem tools as well as reporting issues to PHYSLITE
- Work by ATLAS IRIS-HEP Fellow Sam Kelson ] ] .kol-1-2[
- More on ATLAS Open Data at CHEP 2024:
- The First Release of ATLAS Open Data for Research
(Zach Marshall, Monday plenary) - Open Data at ATLAS: Bringing TeV collisions to the World (Giovanni Guerrieri, Monday Track 8) ]
- The First Release of ATLAS Open Data for Research
.kol-1-2[ .large[
- As columnar analysis .bold[processes events in batches] also need CP tools and algorithms to process in batches
- Current CP tools operate on xAOD event data model (EDM) for calculation and write systematics to disk for future access (I/O heavy)
- Challenge: Can we adapt this model to work in on-the-fly computation columnar paradigm?
- Help with "trade disk for CPU" model
- Refactoring tools to a columnar backend in ATLAS show .bold[improvements in performance and flexibility] ] ] .kol-1-2[
.center[Columnar .cptools[combined performance (CP) tools] operate on .datacolumn[existing columns] in batches to generate .newcolumn[new columns]
(Matthias Vigl, ACAT 2024)]
]
.kol-1-2[ .large[
- Refactoring to columnar CP tools has allowed for .bold[more Pythonic] array interfaces to be developed
- Using next generation of C++/Python binding libraries
 allows
allows
-
Zero-copy operations to/from
$n$ -dimensional array libraries in Python that supports GPUs - Full design control of high-level user API (unified UX) ] ] .kol-1-2[
-
Zero-copy operations to/from
.center[Columnar .cptools[combined performance (CP) tools] operate on .datacolumn[existing columns] in batches to generate .newcolumn[new columns]
(Matthias Vigl, ACAT 2024)]
]
.kol-1-2[
.large[
Demo of prototype (v1) zero-copy Python bindings to columnar Egamma CP tool to compute systematics on the fly for
- Use Uproot to load PHYSLITE Monte Carlo into Awkward arrays
- Apply selections with Uproot and Coffea
- Initialize tools
from atlascp import EgammaTools- Compute systematics on the fly efficiently scaled with dask-awkward on UChicago ATLAS Analysis Facility
] .kol-1-2[
.caption[Selected
(Matthias Vigl, ACAT 2024)]
]
.kol-2-3[ .large[
- v1 prototype established foundations of what was possible with new tooling
- Pythonic interfaces to CP tools could be written without heroic levels of work
- Prototype tools were promising, but more work needed to achieve necessary performance
- No "zero action" option — needed to create standalone prototype to determine if work was reasonable
- v2 prototype takes a step forward in scope
- Moves developments into ATLAS Athena and .bold[migrate ATLAS CP tools to columnar backend] without breaking existing workflows
- Adds thread-safety
- Adds infrastructure support for development of columnar analysis tools
- Allows for full scale integration and performance tests ] ] .kol-1-3[
- Moves developments into ATLAS Athena and .bold[migrate ATLAS CP tools to columnar backend] without breaking existing workflows
]
.huge[
- During (ongoing) refactor added preliminary integrated benchmark to measure .bold[time spent in tool per event] (not I/O) and compare to xAOD model
- While direct comparison not possible, tests are as close as possible
- Only involves
C++CP tool code (no Python involved) - Uses same version of CP tool
- xAOD includes event store access (per-event overhead, paid per-batch in columnar)
- Only involves
- Show .bold[substantial speedups] for migrated tools: .bold[columnar is 2-4x faster] than xAOD interface (EDM access dependent)
- Time for I/O and connecting columns not included in the performance comparisons (not optimized in the tests, so removed from benchmark) ]
.large[
- ATLAS CP tools were created 10-15 years ago to .bold[run in an analysis framework]
- Battle tested, extremely well understood, excellent physics performance, strong desire to be maintained
- Rewrite cost is currently too high across collaboration to move to
correctionlibparadigm - Columnar .bold[cracks open "black box"] implementations of tools for the new analysis model
- Legacy code decisions highlight columnar prototype design decisions and opportunities during tool migration
- Raises the question: "What would it take to get to .bold[
python -m pip install atlascp]?"- Ambitious idea not as far fetched as you might think:
pip install ROOT(Vincenzo Padulano, Monday Track 6)
- Ambitious idea not as far fetched as you might think:
- Columnar prototype explores these possibilities
- .bold[Adopting columnar backend] makes columnar paradigm possible
- .bold[Ongoing
nanobindintegration] bridgesC++/Python with performance - .bold[Pythonic API design] for high level analysis thinking
- Steps beyond: Modularization to level that allows packaging with
scikit-build-core- Allows for "just another" tool in the PyHEP ecosystem ]
.kol-1-2[ .large[
- Tooling ecosystem is proving .bold[approachable and performant] for Pythonic columnar analysis of PHYSLITE
- Enabling mentored university students to implement versions of the AGC by themselves in a Jupyter notebook
- ATLAS IRIS-HEP Fellow Denys Klekots's AGC project using .bold[ATLAS open data] (implementation on GitHub)
- Simplified version of IRIS-HEP AGC top reconstruction challenge using 2015+2016 Run 2 Monte Carlo from the 2024 .bold[ATLAS open data] release ] ] .kol-1-2[ .bold[Event selection]
- 1 charged lepton
-
$\geq 4$ hadronic jets - Lepton kinematics:
$p_{T} \geq 30~\mathrm{GeV}$ ,$\left|\eta\right| < 2.1$ - Jet kinematics:
$p_{T} \geq 25~\mathrm{GeV}$ ,$\left|\eta\right| < 2.4$
.huge[
- Columnar analysis tool efforts inside of ATLAS have been promising with CP tools showing performance increases and bespoke UI
- Development of a columnar ATLAS AGC demonstrator with full systematics is ongoing supported by advancements in v2 prototype
- Technical advancements are being incorporated into ATLAS wide tooling
- Contributions upstream to PyHEP community tools
- ATLAS Open Data proving to be useful for research and community communication
- Advancements in tooling are enabling researchers across career stages ]
.huge[This work was supported in part by the United States National Science Foundation under Cooperative Agreements OAC-1836650 and
PHY-2323298 (Institute for Research and Innovation in Software for High Energy Physics (IRIS-HEP)).]
class: end-slide, center
.huge[Backup]
.center.large.bold[ "columnar analysis" == "array programming for data analysis" ]
.kol-1-2[ .large[
- Higher level APIs for physicists and improved user experience
- People using columnar analysis on ntuples already seem to be loving it
- Enable the same UX but without ntupling (save disk)
- Potential for higher performance
- Enable on-the-fly combined performance (CP) tool corrections on PHYSLITE
- Broader scientific data analysis ecosystem integration
- Extend and scale ATLAS tools with large and performant ecosystem ] ] .kol-1-2[
.center.large[Different expressions/representations for same analysis result goals] .caption[(Nick Smith, 2019 Joint HSF/OSG/WLCG Workshop)] ]
.large.center[ HL-LHC era data scale requires rethinking interacting with data during analysis ]
.kol-2-5[ .large[
- .bold[Analysis Grand Challenge] (AGC) community exercise organized by IRIS-HEP includes the stages of a projected typical HL-LHC analysis
- Demonstrator of development of the required cyberinfrastructure
- The 200Gbps Challenge: Imagining HL-LHC analysis facilities (Alexander Held, Monday plenary)
- Opportunity for ATLAS to demonstrate columnar analysis views and areas for improvement ] ] .kol-3-5[
.center.large[High level view of operations in an HL-LHC analysis] ]
.kol-2-5[
.center.huge[Broader "Scientific Python" ecosystem is designed to be interoperable and support multiple domain levels] ]
.kol-1-5[
].kol-2-5[
.center.huge[Interoperable domain hierarchy design continued in "PyHEP" ecosystem] ]
.kol-1-3[ .large[
- University of Chicago Analysis Facility .bold[provides testing bed] for analysis platform
- Provides support for:
- .bold[JupyterLab] as a common interface
- Highly efficient data delivery with .bold[XCache]
- Conversion to columnar formats with .bold[ServiceX]
- Excellent integration exercise between analysis and operations ] ] .kol-2-3[
.center.large[Scalable platform for interactive (or noninteractive) analysis] ]
.kol-1-2[ .large[
- .bold[First] release of ATLAS Run 2 2015 and 2016 open data in July 2024
- Using ATLAS open data for AGC
- Open access data allows for use in testing community projects and problems
- Released as PHYSLITE (HL-LHC data format)
- Allows for new students to be able to learn analysis and make contributions quickly
- More on ATLAS Open Data at CHEP 2024:
- The First Release of ATLAS Open Data for Research (Zach Marshall, Monday plenary)
- Open Data at ATLAS: Bringing TeV collisions to the World (Giovanni Guerrieri, Monday Track 8) ] ] .kol-1-2[
.large[
- For reading
ElementLinkand other unreadable members are pursuing multiple strategies - Have Awkward behaviors in Python, but we also try to turn everything into "plain old data" (POD) branches, and RNTuple will help with that
- If only target infrastructure was Uproot we could stick with Awkward behaviors, but RDF (without dictionaries), and Julia would also have to support such custom reading, and that's not a scalable approach ]
- ATLAS Software and Computing HL-LHC Roadmap, ATLAS Collaboration, 2022
- Documentation on PHYSLITE Variables for ATLAS Open Data, ATLAS Collaboration, Accessed 2024
- Using Legacy ATLAS C++ Calibration Tools in Modern Columnar Analysis Environments, Matthias Vigl, ACAT 2024
- How the Scientific Python ecosystem helps answering fundamental questions of the Universe, Vangelis Kourlitis, Matthew Feickert, and Gordon Watts, SciPy 2024
- ATLAS PHYSLITE Content Documentation, ATLAS Collaboration, Accessed 2024 [ATLAS Internal]
- The Columnar Analysis Grand Challenge Demonstrator, Gordon Watts, ATLAS S&C Plenary Afternoon: Demonstrators, 2023-10-04 [ATLAS Internal]
- ATLAS AGC Demonstrator, Gordon Watts, ATLAS AMG+ADC Joint Session, 2023-03-30 [ATLAS Internal]
- Tour of the CP Columnar Prototype and CP Algorithm Conversion, Nils Krumnack, 2024-10-07 [ATLAS Internal]