The main objective of this course is to demonstrate how a bioinformatics workflow can be seamlessly run using TACC resources from raw input to biological interpretation. There are many different types of workflows you can run using the systems we have available here at TACC. In particular, transcriptome analysis will be demoed as it is a familiar concept among wide range of disciplines.
- Understand Transcriptome analysis
- Run through a RNA-seq data analysis pipeline using Stampede
Transcriptome is simply defined as all of RNA molecules in a single cell.
- Represents expression levels of all genes for a single condition of cell for a specie at given time.
- Also Rereferred as expression profiling.
- Unlike Genome analysis can show differences in the exact same cell that are in different external environment.
Only a decade ago, the study of gene expression was limited to human genetics for medical purposes or model organisms such as mouse, fruit fly and nematodes. Then, microarrays and serial analyses of gene expression were the only available tools for examining transcriptome. With the recent advances of next-generation sequencing technologies, the cost effectiveness of sequencing and maturation of analytical tools, the transcriptome analysis has become more a realistic option for genetic nonmodel organisms, even for individual laboratories.
Transcriptome studies are often featured in give high-impact journals as it describes status of cells in different conditions in context of expression levels. Here are some recent examples of these in Science, Nature and Cell
The pipeline uses Next-generation sequencing RNA-seq data to as a raw input.
Sequence Read Archive
https://www.ncbi.nlm.nih.gov/sra/SRX2771645
https://www.ncbi.nlm.nih.gov/sra/SRX2771647
Genome index and annotation
https://ccb.jhu.edu/software/tophat/igenomes.shtml
- Hands-on: Try downloading and extracting genome index and annotation files to a directory
wget ftp://igenome:[email protected]/Arabidopsis_thaliana/Ensembl/TAIR10/Arabidopsis_thaliana_Ensembl_TAIR10.tar.gz
tar -zxvf Arabidopsis_thaliana_Ensembl_TAIR10.tar.gz
SRAtoolKit
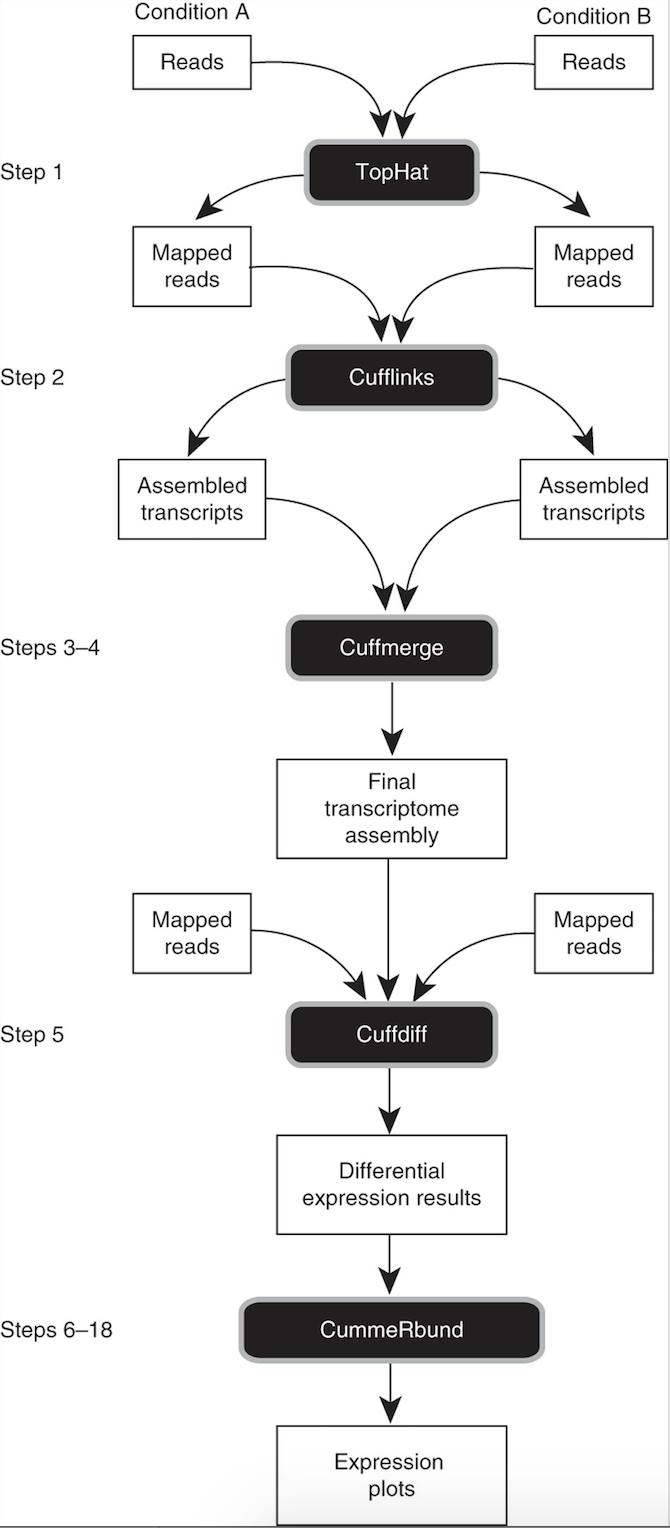
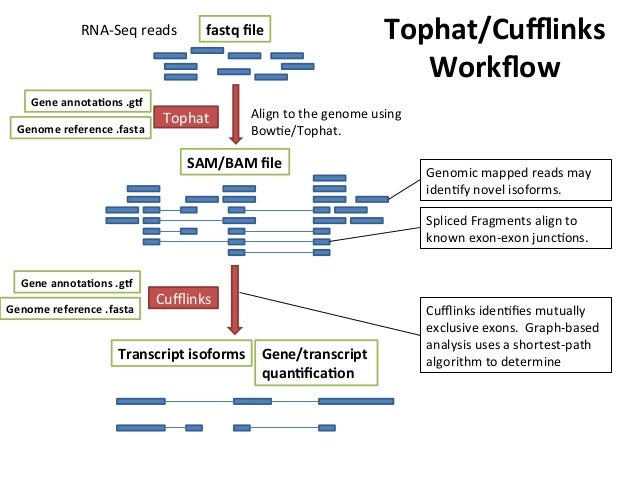
Tuxedo suite: includes: TopHat, Cufflinks, Cuffmerge, Cuffdiff (cite) https://www.nature.com/article-assets/npg/nprot/journal/v7/n3/images_article/nprot.2012.016-F1.jpg
Samtools
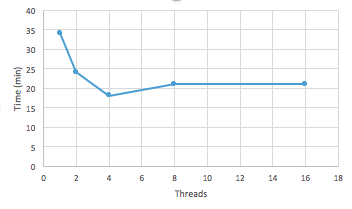
What is the advantage of using TACC vs. workstation on transcriptome analysis? Testing number of threads and CPU time for an alignment to complete
The two sequencing files are of a plant model organism, Arabidopsis Thaliana.
- A member of the Brassica family
- A favorite research subject for plant molecular geneticists
- Due to its remarkably small genome size (about 100,000 kB, with less than 25% repetitive sequences)
- And ease of culture in the lab
- Short stature and life cycle
- Ameanable to genetic mapping studies and other techniques (such as transformation and gene cloning).
-
Col-0 strain, which is columbia seed.
-
A widely used wild type seed, selected for its high fertility, vigor, and responsiveness to changes in photoperiod.
-
Contains no visible genetic markers.
-
Second is of jazQ mutant, which has enhanced JA-regulated defense against insect herbivory without an associated reduction in leaf growth.
-
Is a JA signalling mutant (jazQ), in which the removal of multiple JAZ repressors causes hyperactivation of JA responses
-
As a consequence, jazQ plants exhibit both enhanced resistance to insect herbivory and diminished growth of leaves and roots.
First hands on:
- Convert SRA file to FASTQ
module load sratoolkit
scratch_cache
prefetch SRR5488800 ; fastq-dump SRR5488800
prefetch SRR5488802 ; fastq-dump SRR5488802
cp /scratch/02114/wonaya/SSI/SRR5488800.fastq .
cp /scratch/02114/wonaya/SSI/SRR5488802.fastq .
(2 minutes)
stampede2:
module load bowtie tophat
module load boost on ls5
TopHat run: Aligning sequences on arabidopsis genome guided with gene annotations
tophat2 -p 4 -G Arabidopsis_thaliana/Ensembl/TAIR10/Annotation/Genes/genes.gtf -o test_1 --no-novel-juncs Arabidopsis_thaliana/Ensembl/TAIR10/Sequence/Bowtie2Index/genome SRR5488800_1m.fq
tophat2 -p 4 -G Arabidopsis_thaliana/Ensembl/TAIR10/Annotation/Genes/genes.gtf -o test_2 --no-novel-juncs Arabidopsis_thaliana/Ensembl/TAIR10/Sequence/Bowtie2Index/genome SRR5488802_1m.fq
cp -r /scratch/02114/wonaya/SSI/test_1/ . ; cp -r /scratch/02114/wonaya/SSI/test_2/ .
(15*2 minutes)
Cufflinks:
cufflinks -p 16 -o wt_cuff -G Arabidopsis_thaliana/Ensembl/TAIR10/Annotation/Genes/genes.gtf test_1/accepted_hits.bam
cufflinks -p 16-o sa_cuff -G Arabidopsis_thaliana/Ensembl/TAIR10/Annotation/Genes/genes.gtf test_2/accepted_hits.bam
cp -r /scratch/02114/wonaya/SSI/wt_cuff/ . ; cp -r /scratch/02114/wonaya/SSI/sa_cuff/ .
(1*2 minutes)
Cuffmerge:
cuffmerge -p 16 -g Arabidopsis_thaliana/Ensembl/TAIR10/Annotation/Genes/genes.gtf -s Arabidopsis_thaliana/Ensembl/TAIR10/Sequence/WholeGenomeFasta/genome.fa cuffmerge.txt
in which the content of cuffmerge.txt should be (using text editor such as vim):
wt_cuff/transcripts.gtf
sa_cuff/transcripts.gtf
cp /scratch/02114/wonaya/SSI/cuffmerge.txt .
(1 minutes)
Cuffdiff:
cuffdiff -L WT,SA -p 16 merged_asm/merged.gtf test_1/accepted_hits.bam test_2/accepted_hits.bam
cp -r /scratch/02114/wonaya/SSI/merged_asm .
(11 minutes)
cat test_1/align_summary.txt
How efficient was the alignment? TopHat reports alignment statistics internally, but other tools may not. In these cases, use samtools flagstat
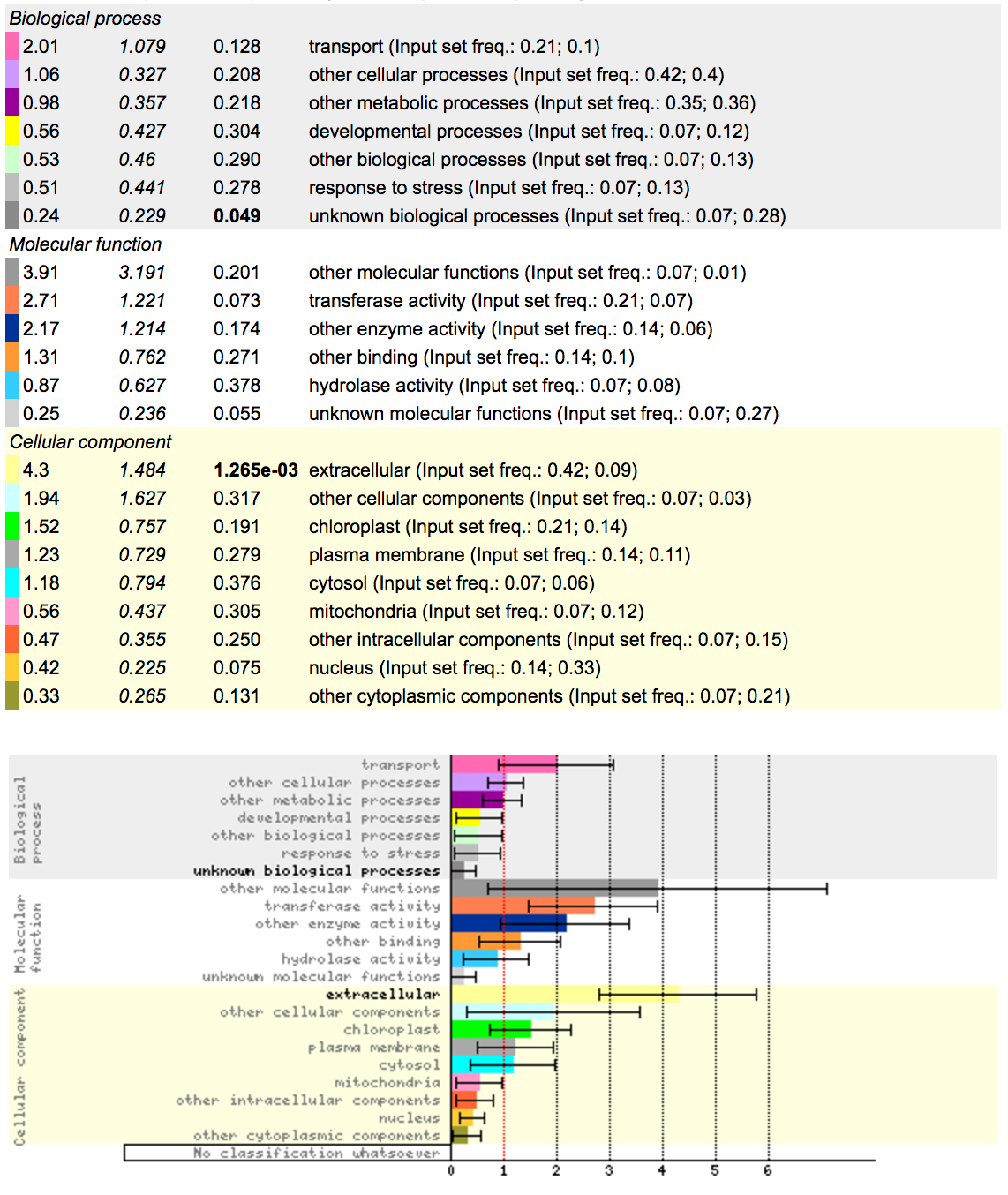
So with this data, what can you deduce?
http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi
Using the dataset you got from running cuffdiff, you should be able to extract top 100 genes with highest log-fold changes in expression. Save the names of these genes as a separate text file, and copy the names of genes to the tool above. What functional enrichment do you see in mutant strain?
sort -nrk 10 gene_exp.diff > gene_exp.diff.sort
head -100 gene_exp.diff.sort > gene_exp.diff.sort.top100
cut -f 3 gene_exp.diff.sort.top100 > gene_exp.diff.sort.top100.names
or Can you pipe this into one process?
sort -nrk 10 gene_exp.diff | head -100 - | cut -f 3 - > gene_exp.diff.sort.nrk.top100.names
Removal of multiple JAZ genes in this jaz quintuple (jazQ) mutant led to constitutive activation of JA responses, causing hypersensitivity to exogenous JA treatment, upregulation of defense-related genes, increased production of secondary metabolites and higher resistance to insect herbivory attack Campos et al. Nat. Comms. 2016 https://www.ncbi.nlm.nih.gov/pubmed/27573094
- Try submitting to TACC system using a job script for tophat to cuffdiff workflow
- Explore different RNAseq dataset
https://www.ncbi.nlm.nih.gov/sra/SRX2834995 https://www.ncbi.nlm.nih.gov/sra/SRX2834994