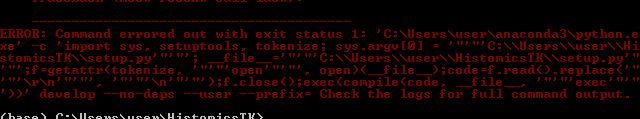
Noob here . I’ve been looking for a python package to work with H&E stained histopathological images. Came upon this. I wanted to install this on Anaconda Environment in Windows 7 .Followed this link Guide to Windows Installation · Issue #855 · DigitalSlideArchive/HistomicsTK · GitHub line by line to install on Windows. But by the time I try to use `python -m pip install -e .’
After running setup.py ,The following error occurs.
Is there any other way to install histomicstk on Windows . I want to use it on an Anaconda environment for my projects.
We don’t test on Windows, and Windows 7 is now considered too old for support. You can run HistomicsTK on Windows 10 in Docker Desktop with WSL enabled, or use Vagrant to get a virtual box that can run things. Otherwise, you probably can install a bunch of libraries and get it working, but we don’t have direct experience doing so.
– David